Lab Automation, Protocol Development and Fast
Reproducible Experiments
Central to good science is the ability to take reproducible measurements in a cost and time effective way. For studies that focus on bacterial physiology and fitness, this means highly accurate and stable growth rate measurements.
Historically, no media or methodology was available for high-throughput and reproducible measurements of the growth rate of aerobic microbes, particularly for the organism I study, Methylobacterium extorquens.
Automated Measurements: I created a unified robotic system to measure and analyze up to thousands of simultaneously growing bacterial populations. Previous investigators have used robotics and microplates to measure the growth rate of cells in microplates (e.g. the Bioscreen product line). Although these systems can create repeated measurements, they do not measure cells under consistent conditions. Oxygen mixing is very poor in the 96, 100 or 384-well plates commonly used leading to stratification, environmental heterogeneity or simply an inability to grow when the metabolism (e.g. methylotrophy) depends on a consistent oxygen supply. Additionally, available products and methods do not scale to more than one plate being measured simultaneously without severe reliability issues.
I fixed these problems by integrating a number of distinct, but off-the-shelf available, components. First, I found a robotic incubator system capable of holding a high volume of plates and a 48-well plate able to provide complete oxygen exchange and mixing (interestingly, many 48 well plates cannot do this). Traditionally, integrating this incubator with a robotic arm, liquid handler, plate reader and other instruments is done by purchasing one of the integration/task-scheduler software packages available from vendors such as PAA or Caliper Life Sciences. However, theses products cost >$5,000 dollars and are usually not suitable for academic science as they are designed to be used in pharmaceutical industries for simple tasks or in prodigal labs where an consultant from the company can be paid to generate implementations or work-arounds for the deficiencies of the original software. I created an open-source modular task scheduler that allowed for arbitrary research designs and equipment integration to circumvent these limitations.
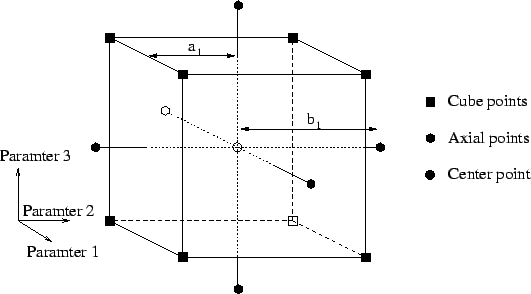
Media Development: I used
response surface methodologies to create a
robust media that also allows for faster growth and experiments that
are reproducible as media ages or different batches are made. Although centrally important to most bacterial studies, little
attention is usually paid to the media used to grow cells.
 Undefined media (e.g. LB) is often used despite causing
differences when experiments are repeated,
and well-defined media can be similarly unreliable as the recipe
often comes from a "first thing that worked" or a "one-factor-at-a-time"
method of optimization. I used a sequential approach
incorporating fractional factorials and central composite designs to
create a media that allows much faster and much more reproducible
growth than what was currently available. The final response
surface was exceptionally flat meaning the media is stable to any
plausibly introduced
variations created by errors or inaccuracies while it is being made
or ageing.
Undefined media (e.g. LB) is often used despite causing
differences when experiments are repeated,
and well-defined media can be similarly unreliable as the recipe
often comes from a "first thing that worked" or a "one-factor-at-a-time"
method of optimization. I used a sequential approach
incorporating fractional factorials and central composite designs to
create a media that allows much faster and much more reproducible
growth than what was currently available. The final response
surface was exceptionally flat meaning the media is stable to any
plausibly introduced
variations created by errors or inaccuracies while it is being made
or ageing.

A biofilm forming in a microplate well.
Strain Modification: By finding and knocking out a gene in biofilm formation, I allowed for meaningful measurements of growth rates by OD to be made in this species. Not all scientifically interesting bacteria that come out of the wild amenable to lab studies. For Methylobacterium an issue in measuring growth curves is that they can stochastically start forming biofilms, leading to a clear violation of beer's law and creates meaningless data. By genetically removing an operon in this species, we were able to fix this issue. I find one of the joys of being trained in both general statistical experimental design and the subject matter whose data you are analyzing is that you can create fundamental changes like this rather than simply optimizing over existing variables.