Epistatic patterns within a gene
Collaborators: Dan Weinreich, Mark Depristo, Daniel Hartl
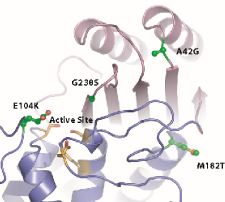
Schematic representation of the Beta-lacatamase protein we studied. The sites I mutagenized are labelled in the diagram.
In my first research project investigating epistatic interactions, we investigated the the type of interactions between advantageous alleles within a protein. We found a general pattern of non-additivity in genetic interactions and because the form these interactions often made the alleles only conditionally beneficial, this greatly reduced the number of mutational trajectories available to evolution, which in turn tends to make recombination less beneficial and evolution much slower. This work was published in Science and is available here.
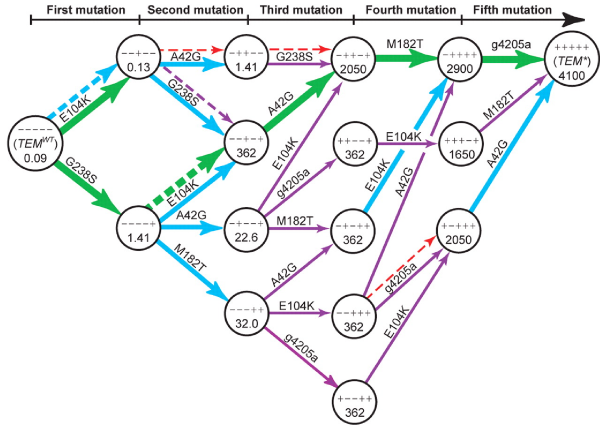
Graphical representation of the portion of the fitness landscape that is selectively accessible. Although 120 mutational trajectories could have been possible, very few of these were because of the epistatic interactions which made most genetic effects only conditionally beneficial.