Epistasis between genes
Collaborators: Hsin-Hung Chou, Hsuan-Chao Chiu, Daniel Segre, Christopher J. Marx
We showed by combinatorial genetics that there is a general pattern of diminishing returns epistasis between loci in different genes. This has important consequences and our paper was recently published in Science (paper here and supplements here). Kryazhimskiy, Draghi and Plotkin also wrote a great perspectives article that is a nice introduction to the work for anyone who hasn't seen it yet.
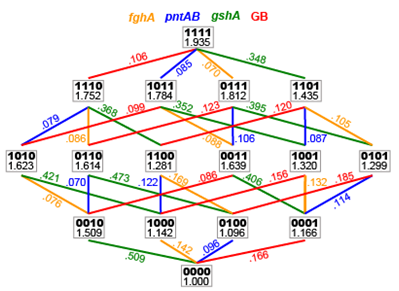
The fitness landscape studied in this project. It was determined by making (almost) all 2N different genetic combinations and assessing the values between them.
This work builds on my earlier work examining epistasis for loci within a gene, and I have also explored the implications of this pattern for genetic associations for anyone who wants to learn more about the importance of epistasis for how biology works and how we study it.

The cell shape phenotype related to fitness in this study.
There is often a lot more to a study than can be fit into a Science paper. A particularly interesting aspect of this data, and one that we did not have room to discuss, is that the model in the paper is more complicated and has less predictive power than a much simpler model. In particular, by simply transforming how we define fitness all epistasis in the system disappears and it is possible to fully predict all 2N phenotypes from only (N+1) observed phenotypes, which ironically means that our paper about epistasis has no epistasis in the data!
However, the form of this simpler model, series expansions to show why it performs similarly to the model we presented, and my argument for our choice to use the more complicated model is available here for those with a deeper interest.